ChromosomePage
6
6
The DNA of prokaryotes is similarly associated with proteins, some of which presumably function as histones do, packing the DNA within the bacterial cell.
Histones, however are unique feature of eukaryotic cells and are responsible for distinct structural organization of eukaryotic chromatin
Slide 35
The basic structural unit of chromatin, the nucleosome, was described by Roger Kornberg in 1974.
Two types of experiments led to Kornberg’s proposal of the nucleosome model.
First, partial digestion of chromatin with micrococcal nuclease (an enzyme that degrades DNA) was found to yield DNA fragments approximately 200 base pairs long.
In contrast, a similar digestion of naked DNA (not associated with protein) yielded a continuous smear randomly sized fragments.
These results suggest that the binding of proteins to DNA in chromatin protects the regions of DNA from
nuclease digestion, so that enzyme can
attack DNA only at sites separated by
approximately 200 base pairs.
Slide 36
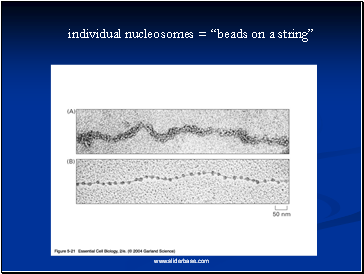
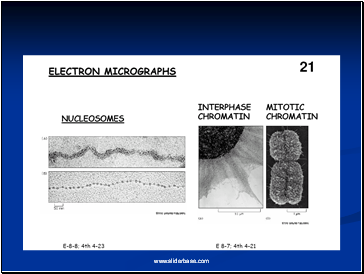
Electron microscopy revealed that chromatin fibers have a beaded appearance, with the beads spaced at intervals of approximately 200 base pairs.
Thus, both nuclease digestion and the electron microscopic studies suggest that chromatin is composed of repeating 200 base pair unit, which were called nucleosome.
Slide 37
individual nucleosomes = “beads on a string”
Slide 38
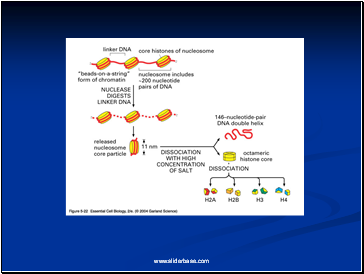
Detailed analysis of these nucleosome core particles has shown that they contain 146 base pairs of DNA wrapped 1.75 times around a histone core consisting of two molecules each of H2A, H2B, H3, and H4 (the core histones).
One molecule of the fifth histone H1, is bound to the DNA as it enters and exists each nucleosome core particle.
This forms a chromatin subunit known as chromatosome, which consist of 166 base pairs of DNA wrapped around histone core and held in place by H1 (a linker histone)
Slide 39
Slide 40
Slide 41
Slide 42
Slide 43
Slide 44
Slide 45
Centromeres and Telomeres
Contents
- What Exactly is a chromosome?
- Number of chromosomes
- Chromosome Size
- Euchromatin and Heterochromatin
- Satellite DNAs
- Prokaryotic and Eukaryotic Chromosomes
- Prokaryotic chromosome
- Bacterial Chromosome
- Supercoiling
- Mechanism of folding of a bacterial chromosome
- Chromatin
- Centromeres and Telomeres
- Centromere
- Kinetochore
- Telomere
- Telomere Repeat Sequences
- Staining and Banding chromosome
- Chromosomal Aberrations
- Structural Chromosomal Aberrations
- Deletion or deficiency
- Deletion in Prokaryotes
- Duplication
- Origin
- Inversion
- Inversions in natural populations
- Translocation
- Non-Disjunction
- Variation in chromosome number
- More about Aneuploidy
- Uses of Aneuploidy
- Trisomy in Humans
- Amniocentesis for Detecting Aneuploidy
- Other Syndromes
- Giant chromosomes
- Lampbrush Chromosome
- Dosage Compensation
- Barr Bodies
- Mechanism of X-chromosome Inactivation
- Reading assignment
Last added presentations
- Sound
- Radiation
- Static and Kinetic Friction
- Motion
- Radioactivity and Nuclear Reactions
- Newton’s law of universal gravitation
- Sensory and Motor Mechanisms