PCRPage
1
1
Slide 1
Polymerase Chain Reaction
Aims
To understand the process of PCR and its uses.
Starter - Match each term with its correct description (work in pairs)
04 September 2015
Slide 2
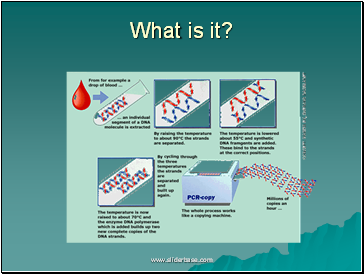
What is it?
Slide 3
Animations
http://www.dnalc.org/ddnalc/resources/pcr.html
http://www.maxanim.com/genetics/PCR/pcr.swf
Slide 4
Slide 5
Slide 6
Information
Polymerase chain reaction enables large amounts of DNA to be produced from very small samples (0.1ml)
There is a repeating cycle of:
separation of double DNA strands
synthesis of a complementary strand for each
Slide 7
What happens?
Sample DNA , nucleotides, DNA primers & thermostable DNA polymerase placed in PCR machine.
Strands of sample DNA separated by heating to 95oC
Mixture cooled to 37oC to allow primers to bind.
Mixture heated to 72oC for replication (optimum temp of DNA polymerase)
Cycle repeats many times (~8mins /cycle)
Slide 8
Problems
Separation achieved by heating to 95oC – no suitable helicase
DNA polymerase can’t work on completely single stranded DNA – double stranded regions needed at the start of sequence to be copied:
primers (short sequences DNA) complementary to bases at start of region to be copied used
To synthesize primers , base sequence at start must be known
Slide 9
TTAACGGGGCCCTTTAAA .…TTTAAACCCGGGTTT
AATTGCCCCGGGAAATTT .…AAATTTGGGCCCAAA
the sequence of bases which ONLY flank a particular region of a particular organism's DNA, and NO OTHER ORGANISM'S DNA. This region would be a target sequence for PCR.
Slide 10
The first step for PCR would be to synthesize "primers" of about 20 letters-long
ONE primer exactly like the lower left-hand sequence, and ONE primer exactly like the upper right-hand sequence:
TTAACGGGGCCCTTTAAA .…TTTAAACCCGGGTT
AATTGCCCCGGGAAATTT .>
and:
< TTTAAACCCGGGTTT
AATTGCCCCGGGAAATTT AAATTTGGGCCCAAA
Slide 11
1 2
Contents
Last added presentations
- Gravitation
- Heat-Energy on the Move
- Upcoming Classes
- Motion
- Radiation
- Waves & Sound
- History of Modern Astronomy